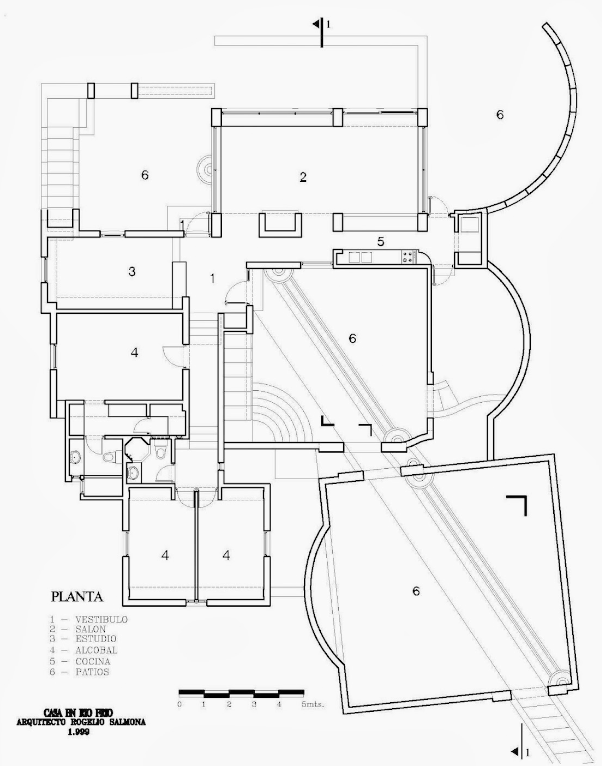
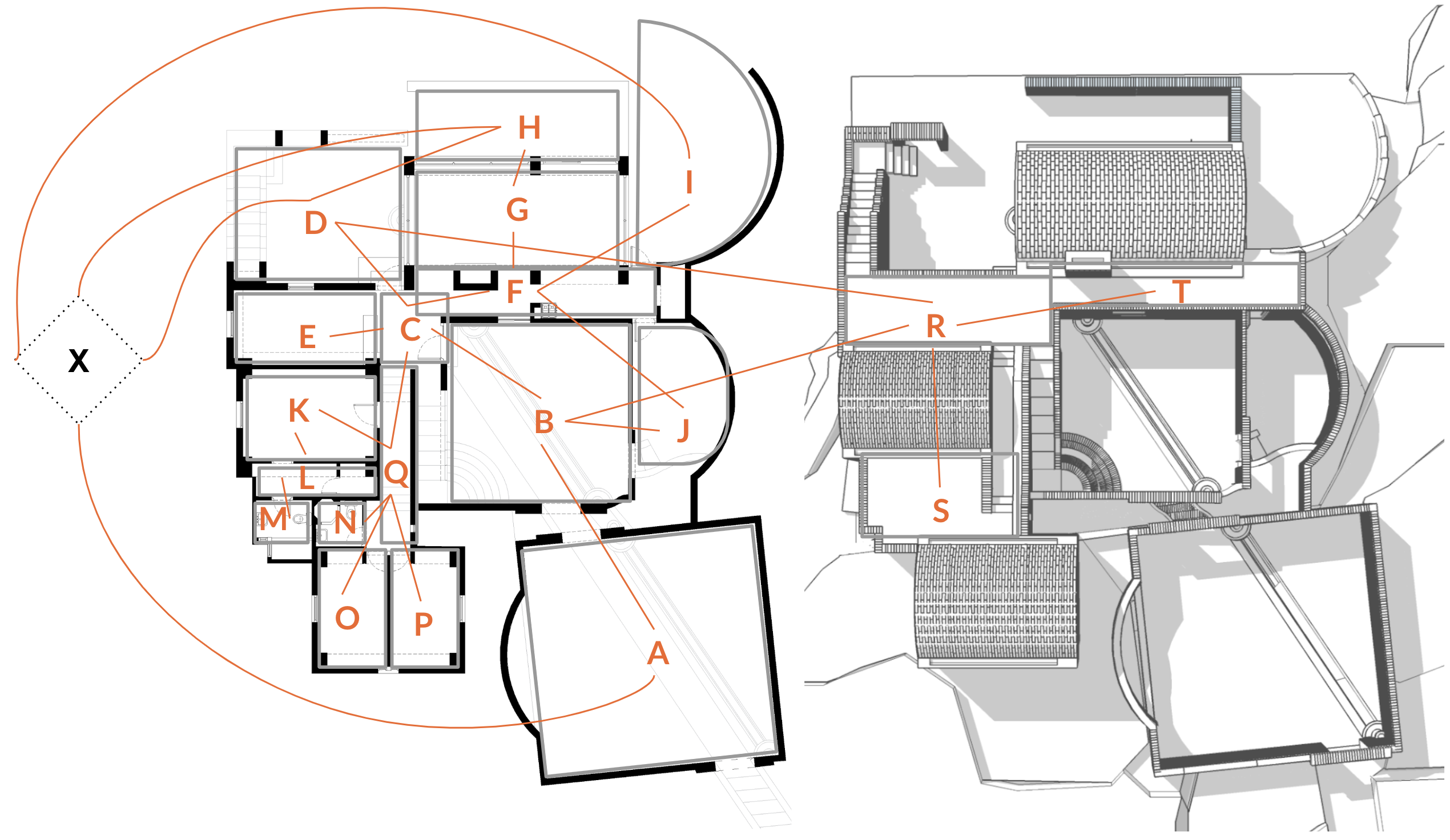
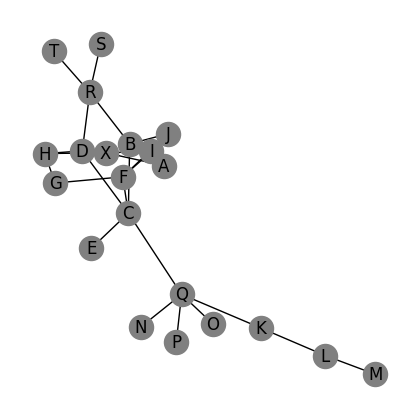
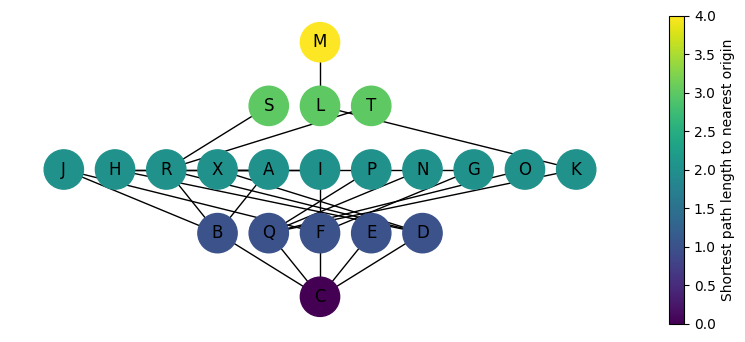
Code
import networkx as nx
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
def jgraph_from_multiple_origins(G, origins, fig_size=(10, 4)):
"""
Draws a Justified graph (J-graph) from multiple origins with a gradient color range.
Handles both a list of origins or a single origin by treating it as a list.
Parameters:
G (networkx.Graph): The graph to visualize.
origins: A single origin or a list of origin nodes for path calculations.
fig_size (tuple): The dimensions for the figure size (width, height).
Returns:
None: Displays the graph visualization with a gradient color scale.
"""
# Ensure origins is a list even if a single node is provided
if not isinstance(origins, list):
origins = [origins]
# Initialize dictionary to keep the shortest paths from any origins
min_path_lengths = {}
# Calculate shortest paths from each origin and update the minimum path length
for origin in origins:
path_lengths = nx.single_source_shortest_path_length(G, source=origin)
for node, length in path_lengths.items():
if node in min_path_lengths:
min_path_lengths[node] = min(min_path_lengths[node], length)
else:
min_path_lengths[node] = length
# Create a new graph to visualize
G_vis = nx.Graph()
# Adding nodes with minimum path length as attribute
for node, length in min_path_lengths.items():
G_vis.add_node(node, steps=length)
# Preserve original edges
G_vis.add_edges_from(G.edges())
# Use a colormap and normalize based on the max path length for color mapping
max_depth = max(min_path_lengths.values())
norm = mcolors.Normalize(vmin=0, vmax=max_depth)
cmap = plt.cm.viridis
# Prepare color values according to the normalized data
colors = [cmap(norm(min_path_lengths[node])) for node in G_vis.nodes()]
# Positioning nodes by topological steps
pos = nx.multipartite_layout(
G_vis, subset_key="steps", align="horizontal", scale=1.75
)
# Drawing the graph
fig, ax = plt.subplots(figsize=fig_size)
nx.draw(G_vis, pos=pos, node_color=colors, with_labels=True, node_size=800, ax=ax)
# Adding a colorbar with correct axis context
sm = plt.cm.ScalarMappable(cmap=cmap, norm=norm)
sm.set_array([])
plt.colorbar(sm, ax=ax, label="Shortest path length to nearest origin")
# Calculating and displaying graph metrics
mean_depth = sum(min_path_lengths.values()) / len(min_path_lengths)
print(f"Mean depth = {mean_depth:.2f}")
print(f"Maximum topological steps = {max_depth}")